Picture Difference in Height Between Hybridized and Ancient Wheat
Abstract
Synthetic hexaploid (SH) wheat (AABBD'D') is developed by artificially generating a fertile hybrid between tetraploid durum wheat (Triticum turgidum, AABB) and diploid wild goat grass (Aegilops tauschii, D'D'). Over three decades, the International Maize and Wheat Improvement Center (CIMMYT) has developed and utilized SH wheat to bridge gene transfer from Ae. tauschii and durum wheat to hexaploid bread wheat. This is a unique example of success utilizing wild relatives in mainstream breeding at large scale worldwide. Our study aimed to determine the genetic contribution of SH wheat to CIMMYT's global spring bread wheat breeding program. We estimated the theoretical and empirical contribution of D' to synthetic derivative lines using the ancestral pedigree and marker information using over 1,600 advanced lines and their parents. The average marker-estimated D' contribution was 17.5% with difference in genome segments suggesting application of differential selection pressure. The pedigree-based contribution was correlated with marker-based estimates without providing chromosome segment specific variation. Results from international yield trials showed that 20% of the lines were synthetic derived with an average D' contribution of 15.6%. Our results underline the importance of SH wheat in maintaining and enhancing genetic diversity and genetic gain over years and is important for development of a more targeted introgression strategy. The study provides retrospective view into development and utilization of SH in the CIMMYT Global Wheat Program.
Introduction
Bread wheat (Triticum aestivum L.; hexaploid genome = AABBDD) naturally evolved via natural hybridization between wild goat grass Aegilops tauschii (DD) and a cultivated emmer plant T. turgidum L. ssp. dicoccon (Schrank) Thell. (2n = 28; AABB, a progenitor of modern durum wheat) around 8,000 years ago1,2. Thus, it consists of three diploid progenitor genomes, AA from Triticum urartu, BB from an unknown species (suggested to be section Sitopsis to which Aegilops speltoides belongs), and DD from Ae.tauschii 3,4. The current form of the Ae. tauschii genome (denoted here as D') is expected to be similar to the original progenitor genome (denoted as D) that led to development of bread wheat. Studies have also suggested that the D genome originated from a homoploid ancestor (derived from the hybridization between A and B diploid progenitors) about five million years ago5,6. The current bread wheat D genome has limited genetic diversity due to (a) hexaploid wheat is expected to have evolved from a few hybridization events with Ae. tauschii (b) there has been limited gene flow from Ae. tauschii to bread wheat, both naturally being highly self-pollinated species with inbreeding coefficients ≥90% and (c) intense human selection of bread wheat led to further depletion of diversity7.
With the development of next-generation sequencing technologies, high-density genome profiling of plant material has become feasible and relatively cost-effective8,9. These new genotyping technologies have already been effectively used to characterize the genetic diversity in bread wheat10,11,12. The limited genetic diversity of the D genome in bread wheat is always reflected by less SNP polymorphisms observed when compared to A and B genomes10,13,14,15. The D' genome is assumed to be structurally similar to the D genome, however as it has not gone through the same extent of natural and human selection, which results in higher SNP diversity16.
Although the introgression of D' genome via SH wheat to synthetic derivative lines can introduce novel variation for traits of interest17,18,19, it may contain unfavorable or detrimental genetic load with complex genetics of pleiotropy and epistatis20. The undesirable component of D' can be reduced by recurrent back or top crossing with elite germplasm and selection. However, in the process of retaining the desirable D' alleles, undesirable alleles at closely linked loci may still be present in synthetic derived lines, causing what is also called 'linkage drag' or 'genetic load'21.
Similar to the D genome, nucleotide diversity in the A and B genomes of bread wheat is also substantially reduced compared to their ancestral progenitors22. The A and B genomes from bread wheat do not reflect or represent the genetic diversity existing in durum wheat23,24. However, bread and durum wheat being both cultivated and selected over generations, the contribution from the durum A and B genomes when introgressed into synthetic derivative lines are not considered detrimental. Thus, SH wheat (AABBD'D') can boost the genetic diversity in all three genomes, as has been well documented25,26,27.
Ae. tauschii harbors substantial variation for many biotic and abiotic stress tolerance traits that are relevant in wheat breeding28. The first attempts to reproduce the bread wheat original crosses were made in the 1940s in Japan29 and US30,31. These attempts led to the development of the first SH wheat28. To upscale impact globally, CIMMYT started to explore the value of wide crosses and the development of SH wheat to increase D genome diversity in the 1980s. The artificial re-creation of bread wheat from improved tetraploid durum wheat and accessions of Ae. tauschii was tested and deployed at larger scale32. The use of improved tetraploid wheat was important to success as two out of the three genomes have already been selected for desirable traits. A cross with wild tetraploid wheat species usually leads to tall synthetics with very undesirable agronomic properties.
When desirable alleles for a trait of interest are considered limited in the current elite germplasm pool, SH wheat is one of first materials to be additionally evaluated in the CIMMYT Global Wheat Program and other bread wheat breeding programs28,33. Synthetic hexaploid wheat have provided valuable diversity for traits related to agronomic and physiological features33,34,35,36,37,38,39, abiotic stress tolerance33,36,40,41,42,43, biotic stress resistance33,35,41,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60, and grain quality61,62,63. To date, the CIMMYT wide-crosses program has developed more than 1,524 SH wheat since the 1980s and has generated thousands of crosses with bread wheat33. Synthetic derivative lines have been selected as parents for mainstream breeding, with rigorous selection resulting in advanced lines with excellent performance for yield and other traits. Synthetic derivative lines have been selected as candidates in international yield trials, which are disseminated every year globally. Over 80 synthetic derivative lines have also been released as cultivars and are widely grown64,65. Linkage drag may discourage wheat breeders to use SH wheat in their breeding programs21. Pre-breeding efforts are required to retain the desirable variability of the D', A and B genomes and reduce the undesirable genetic load. Studies on the genetic contribution of the durum A and B genomes and Ae. tauschii D' genome diversity that remain in synthetic derivatives used and retained in a breeding program are limited. The objectives of our study were to: (a) estimate the genetic contribution of the D' genome via DartSeq® markers in a set of selected synthetic derivative lines and determine its correlation with the expected theoretical contribution calculated using ancestral pedigree information, (b) test if the genetic contribution is disproportionate in different parts of the genome, and (c) measure the theoretical contribution of SH wheat within the best CIMMYT advanced breeding lines distributed in international yield trials over the last 19 years.
Results
Marker-estimated contribution vs. the theoretical contribution of the D' genome in the synthetic derivative lines
After applying the various data filters (see methods), a total of 2,669 D genome specific markers (975 PAV and 1,694 SNPs) were recovered for further analysis (Supplementary Figs S1, S2). The frequency of a D genome specific marker, with a differential frequency between the synthetic hexaploid wheat (shw) and bread wheat (bw) populations, was significantly higher than the A and B genome specific markers, with differential frequencies between the durum wheat (dw) and bw populations (Supplementary Fig. S2). The population allele frequency difference was set to 0.3 falls in 80 percentiles of the distribution. Therefore, only the D genome specific markers were used to estimate of D' contribution from Ae. tauschii to the synthetic derivative lines. The number of differential DarSeq® markers on the A and B genome was considered to be too low to precisely estimate the contribution of the A and B genomes of durum wheat to the synthetic derivative lines.
The average theoretical contribution of the D' genome to the 253 synthetic derivatives in our study was 25.3% (Fig. 1). In contrast the marker-estimated contribution of the D' genome was 17.4% (Fig. 2). Thus, the marker-based estimate was 7.9% lower than the theoretical estimate. However, there was a high correlation between both estimates (r = 0.88**) (Supplementary Fig. S3). The majority of the synthetic derivative lines showed a marker-estimated contribution,, less than expected. The synthetic derivative lines of generation 1 and 2 had on an average a lower D' contribution (ϕ) compared to expectation (φ = 12.5 to 25.0%). On the other hand, synthetic derivative lines of subsequent introgression cycle maintained a somewhat higher D' contribution than expected (Figs 1, 2, Supplementary Fig. S3). The results indicate a relatively rapid selection against the undesirable genome segments of Ae. tauschii during the first introgression cycles but a residual D' contribution due to linkage drag in later introgression cycles.
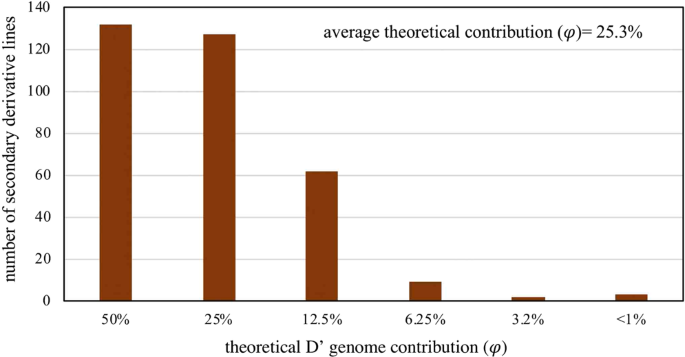
Theoretical D' genome contribution in a set of 253 selected synthetic derivative lines (SD).
Full size image
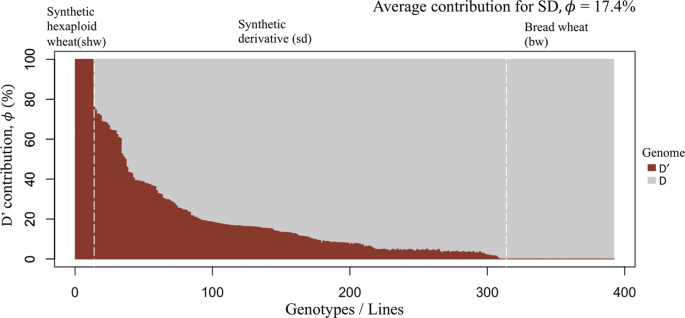
Marker-based estimate of the contribution of D' genome of Ae. tauschii in a set of 253 selected synthetic derivative lines (SD).
Full size image
In a second step, we looked how the contribution of D' genome to the synthetic derivative lines is distributed across the genome (Fig. 3). The average contribution of D' in each chromosome varied, while chromosome 2D was the least contributor and chromosome 7D the highest contributor (Fig. 3). For the majority of chromosome regions, the proportion of the D' versus the D genome contribution was less than 18%. However, some chromosome regions were clearly more likely to have D' alleles over D with a proportion equal to or greater than 50% (Fig. 3). The Supplementary Table S2 provides list of genes retrieved form RefSeqV1.166 in regions with a higher likelihood to retain D' genome alleles.
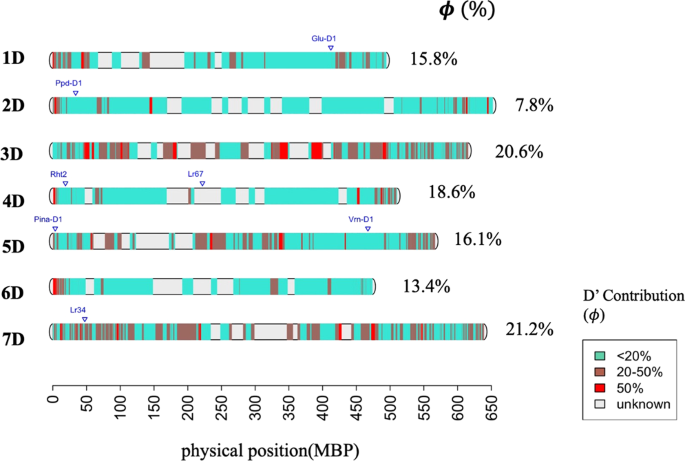
Most likely genomic regions with a D' genome contribution. Physical positions are based in Chinese spring wheat genome sequence RefSeq v1.0.
Full size image
Average genetic contribution to international nursery lines and released cultivars
We also calculated the theoretical contribution of D' in two international yield trials and in released cultivars with SH wheat in its pedigree. The average contribution of the D' genome in synthetic derivative lines was generally higher in the Semi-Arid Wheat Yield Trial (SAWYT) than in the Elite Spring Wheat Yield Trial (ESWYT) (Fig. 4). In both trials, increasing number of synthetic derivatives with yearly fluctuations were retained in recent years. However, the contribution from the D' genome decreased in recent years indicating that subsequent crosses have reduced the D' genome contribution only retaining the favorable D' regions in the genome. Across both international yield trials, 25 SH wheat was successfully utilized in developing competitive synthetic derivatives (Table 1). The SH lines 'ALTAR 84/AEGILOPS SQUARROSA (TAUS)' and 'CROC_1/AEGILOPS SQUARROSA (224)' were the most frequent SH wheat used. The synthetic derived lines, such as SOKOLL (PASTOR/3/ALTAR 84/AEGILOPS SQUARROSA (TAUS)//OPATA) and VOROBEY (CROC_1/AEGILOPS SQUARROSA (224)//OPATA/3/PASTOR) were subsequently used as parents.
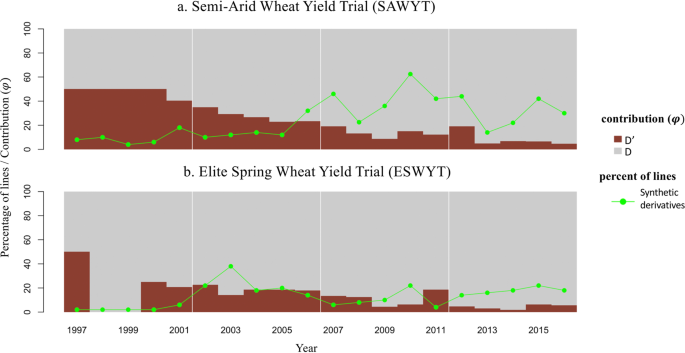
Theoretical contribution, φ and number synthetic derivative lines in two CIMMYT international yield trials SAWYT and ESWYT.
Full size image
Full size table
Among known released cultivars with synthetic background, the released cultivars retained an average of 17.48% of D' genome contribution (Table 2). Noteworthy, cultivars with as high as 50% of D' contribution (synthetic derivative lines of generation 1) have been released, although the majority of cultivars had less than 12.5% of D' contribution (Supplementary Table S2).
Full size table
Discussion
Our study provides a unique perspective of the genetic contribution of Ae. tauschii to the CIMMYT spring bread wheat breeding program via SH wheat, which have been of interest for many years23. Studies have shown that the D genome diversity of SH wheat is considerably higher than of bread wheat25,27. Although direct hybridization of bread wheat with T. turgidum and Ae. tauschii is possible, it is generally difficult to generate and maintain stable genomes67. As a result, the development of SH wheat is considered a better alternative. In addition, SH wheat can also serve as bridge to introduce alleles from tetraploid durum wheat and emmer wheat to hexaploid bread wheat33. In contrast to initial years when few elite durum wheat genotypes were used to develop SH, in more recent years additional lines from the tetraploid Triticum dicoccoides and Triticum dicoccum 20,23,68 pools were introgressed.
The potential of wild relatives of wheat including Ae. tauschii to improve disease resistance in bread wheat is well documented. For example, the leaf rust resistance gene Lr21 was introgressed into the wheat cultivar Thatcher from Ae. tauschii accession TA1599 via SH wheat69. Similarly, gene Yr28 and other genes of resistance to stripe rust have been transferred from Ae. tauschii 70. More recently, two Ug99 stem rust resistance genes (SrTA10187 and SrTA10171) were transferred from Ae. tauschii into the hard-white winter wheat line KS05HW1471. In several instances, synthetic derivative lines have also been reported to positively contribute to yield and abiotic stress torelance19,72,73,74.
Beside Ae. tauschii, other wild wheat relative species can be introgressed into wheat without development of synthetics. However, usually the transfer of the alien segments is practically and methodologically more challenging and creates a situation where no homologous pairing occurs. The D' genome being the progenitor of the D genome, problems of homologous paring have not been reported. Due to its allo-hexaploid nature, bread wheat is in general have high buffering capacity to alien transfers.
The A and B genomes are more closely related with each other than the A vs. D genome or the B vs. D genome5,6. This makes it more difficult to find SNP markers specific to the A or B genome. The A and B genomes from durum and bread wheat have common ancestors, while natural hybridization between the genomes occurred 8,000 years ago. Both genomes have gone through natural and artificial selection over years, thus likely evolved in a similar manner. Moreover, the dwarfing gene Rht1 in modern durum wheat was transferred to bread wheat further allowing the mixing of the two species. Our study clearly showed that the differences between the A or B genomes from durum and bread wheat were smaller than between the D' and D genomes.
The theoretical contribution of the D' genome was a good predictor of the estimated marker-estimated D' genome contribution. However, synthetic derivatives within each theoretical contribution class (breeding cycles 1 to 7) differed significantly for the estimated marker- contribution and for genomic regions deriving from the D' genome. Thus, the theoretical estimates provide some indication about the potential contribution of D', but is not an accurate reflection due to natural and artificial selection, linkage drag and Mendelian sampling that occurs during the breeding process.
Adaptive introgression is very common phenomena, which leads to unbalanced contributions in different part of the genome particularly in regions vital for plant survival and adoption75. SH wheat shows a number of undesirable traits such as a shattering rachis, glume retention, glume hardiness and tall plant height33,67. Therefore, during the process of developing synthetic derivatives, large progeny populations (exceeding 2,000 to 3,000 plants in size) were grown to select for agronomically desirable traits particularly threshability, plant height and disease resistance33. Natural adaption and adaptive introgression apply as some progenies do not survive due to chlorosis or necrosis. In addition, the fitness advantage of wild or domesticate alleles may be favored in process of natural selection76,77,78.
The large progeny populations are used by breeders for stringent selection. The probability of retaining D' is higher in regions surrounding QTL/genes of interest when certain plant types and target traits are selected and can last several generations of backcrossing21. Our results showed a larger extent of the residual D' contribution was possibly due to linkage drag in later breeding cycles. We also demonstrated that there is no clear biased preference among alleles in the majority of genome regions. However, some bias in contribution is obvious as during the process of developing the synthetic derivatives both natural and artificial selection are applied, possibly favoring genes related to survival and to typical bread wheat plant types. The search for the underlying genes in the preferable selected D' genome segments did not provide any deeper insight as the function of most genes was unknown.
CIMMYT established its Wide Crosses Program, in 1986. Since their development, synthetic derivative lines were then frequently used in mainstream breeding and the superior progenies were selected for inclusion and distribution through international yield trials and nurseries, a few were released as cultivars. In the past 32 years, CIMMYT has generated more than 1,401 spring type SH wheat and over two thousand crosses made between the most promising SH wheat and elite bread wheat lines. Generating winter type SH wheat started in 2008. Approximately 50 targeted SH wheat are currently developed by CIMMYT annually, by crossing current elite durum wheat cultivars, but extending the genetic diversity of the Ae. tauschii accessions by using combined genotypic and phenotypic information.
The number of SH wheat increased in both yield trials (ESWYT and SAWYT) during the last 19 years, however there was a declining trend of the D' genome contribution on an individual basis. Synthetic derivatives of breeding cycle 1 with a D' genome contribution of 50% formed part in yield trials disseminated from 1997 to 2000, whereas in more recent years the average D' genome contribution was less than 15%. Although 1524 SH wheat were developed to date, derivatives from 25 SH wheat reached the yield trials. Over one thousand initially developed SH wheat were characterized for agronomic and other economically important traits and below 10% were found to have potential for use in breeding programs; the set is available as Elite Synthetics from the CIMMYT germplasm bank. Similarly, thousands of synthetic derivatives have been characterized over the years, some now released as varieties, however the most popular synthetic derivatives, e.g. SOKOLL and VOROBEY, were used multiple times as parents in mainstream breeding due to their superior performance under drought stress and resistance to Septoria tritici blight. The SH wheat ALTAR_84/AEGILOPS SQUARROSA(TAUS) was the most frequent line in both ESWYT and SAWYT. Among the durum wheat donors ALTAR_84 dominated followed by CROC_1 (=LAHN) and CHEN. ALTAR_84 was bred during the 1980s based on a new ideotype concept, with balanced increase in all yield components and was extensively grown in Mexico. The higher percentage of SH wheat in the SAWYT is due to their potential introgression of new stress tolerance genes79. A more recent outcome of the utilization of SH has been the development and release of high grain Zn, biofortified, varieties 'Zn Shakti' from a durum wheat SH, and 'WB-02', 'PBWZn-1′, and 'Nohely F2018' from T. dicoccum based SH wheat.
The value of introgression of SH wheat is also demonstrated by an increasing number of varieties released in various countries. It is difficult to estimate the use of synthetic derivative lines as parents across breeding programs due to the (a) unavailability of pedigree record of cultivars released in different countries and (b) incomplete or inaccurate pedigree records available. Based on available pedigree records some cultivars are released with as high as 50% theoretical D' genome contribution (e.g. KT2009 in Pakistan). This means the buffering capacity of the A and B genome can mask undesirable effects of wild alleles.
The development of SH wheat led to increased D genome diversity via the introgression of D' at CIMMYT. However, for SH wheat to be increasingly used in mainstream breeding, a targeted introgression of high value traits combined with a proper selection strategy through limited crossing is considered important to capture additional useful genes/diversity from SHs. The breeding strategy needs to consider minimizing the unfavorable contribution of the D' genome due to linkage drag while maintaining the desirable alleles. This raises the question, how much of D' genome contribution is acceptable or favorable in synthetic derivatives or what is the average percentage of D' genome contribution that must be discarded using selection. The theoretical contribution in the most recent CIMMYT international yield trials remained less than 12.5% contribution of the D' genome which means that at least three rounds of crosses with elite bread wheat lines is required if backcrossing and stringent selection for multiple traits is not applied. As we have seen the difference between the theoretical and marker-estimated contribution of D' at the individual and across genome level, accelerating the process through an appropriate breeding approach can only be archived by getting rid of the unnecessary genome segments of D' while retaining desirable genes/QTLs. Combining genomic profiles with the phenotypic assessment of SH wheat could help to identify the desirable genes/QTL, which could be traced during the selection process via genomic-assisted breeding approaches.
Conclusion
The results show that the genetic contribution of the D' genome of Ae. tauschii can be estimated using high-density genome profiles. The marker-estimated contribution of D' in this study underlines the importance of SH wheat in maintaining diversity and genetic gain over years. However, targeted breeding efforts are required to effectively use the D' diversity for mining desirable alleles. Molecular markers, if tagged to desirable alleles, will have the potential to accelerate the selection process to maintain the desirable variation while culling the undesirable variation. Overall, the development and utilization of SH wheat at large scale at CIMMYT is a model for maintaining diversity in bread wheat breeding germplasm required for combating future challenges in crop improvement and is a successful example of the utilization of genetic diversity from wild relatives.
Methods
The SH wheat included in this study were developed by CIMMYT wide-crosses program by crossing durum or emmer wheat (Triticum dicoccum, AABB) with Ae. tauschii (D'D'), followed by artificial doubling of the chromosomes of resulting haploid F1 plants (Fig. 5, Supplementary Fig. S5). The SH wheat was then crossed with bread wheat lines via 1 or 2 backcrosses or 3-way (top) crosses, to generate advanced lines through selections conducted in segregating generations and as fixed lines; known as a synthetic derivative line (Supplementary Fig. S5). The SH wheat crossed "x" times with bread wheat is termed as synthetic derivative line of breeding cycle "x". For example, synthetic derivative line of breeding cycle 1 is defined by an SH wheat crossed once with a bread wheat.
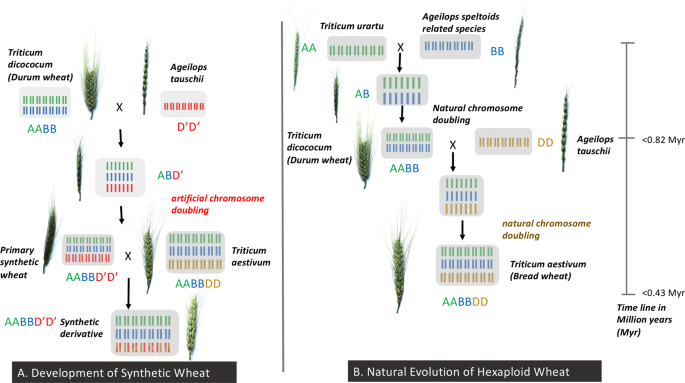
Development of synthetic hexaploid wheat (AABBDD') in comparison to the emulating evolution of the hexaploid wheat (AABBDD).
Full size image
Marker-based contribution estimation
A total of 359 genotypes used in this study included three Ae. tauschii lines, 30 durum wheat lines, eight SH wheat, 253 synthetic derivative lines, and 63 bread wheat lines (Supplementary Table S3). Synthetic derivative lines were between 1 to 7 cycles in derivation. The synthetic derivative lines were selected because of their relevance in the CIMMYT Spring Bread Wheat Improvement Program, with at least one sister line distributed through CIMMYT international yield trials, but with at least 3 to 4 sister lines conserved in the CIMMYT Wheat Germplasm Bank.
All entries were genotyped with the DArTseq® technology at the Genetic Analysis Service for Agriculture (SAGA) laboratory at CIMMYT in Mexico. A complexity reduction method including two enzymes (PstI and HpaII) was used to create a genome representation of the set of samples. A PstI-RE site specific adapter was tagged with 96 different barcodes enabling multiplexing a 96-well microtiter plate with equimolar amounts of amplification products within a single lane on an Illumina HiSeq2500 instrument (Illumina Inc., San Diego, CA). The successful amplified fragments were sequenced up to 77 bases, generating approximately 500,000 unique reads per sample. A proprietary analytical pipeline developed by DArT P/L was used to generate allele calls for SNP and SilicoDArT. Then, a set of filtering parameter was applied to select high quality markers for this specific study. A total 76,543 Single Nucleotide Polymorphism (SNPs) and 365,663 SilicoDArT were revealed. SilicoDArT markers are a secondary marker type provided by DArTseq®, which are scored for the presence or absence of a single loci. The genotypic data were then filtered based on the following consecutive steps:
- (a)
Markers with known chromosome position on the Chinese Spring reference genome sequence RefSeq v1.066 (number of markers retained = 211,169). Markers with unknown genomic position were removed.
- (b)
Markers genome specific for the A, B or D genomes, respectively (number of markers retained = 202,668).
- (c)
Markers with a missing data rate less than 30% (number of markers retained = 188,584)
- (d)
Markers with minor allele frequency MAF ≥ 0.05 and heterozygote frequency HET ≤ 0.1 (number of markers retained = 37,201)
- (e)
Markers with a population specific allele differential greater than 0.30 (see equation 1 and 2, below) (number of markers retained for D genome = 2,669). Such arbitrary threshold was set to reduce number of markers that are most informative. To determine a marker-estimated genetic contribution of the D' genome in the selected set of synthetic derivative lines, we only used markers with a large allele-frequency difference among the populations, i.e. markers with population-specific alleles80,81.
The primary populations in our study included the Ae. tauschii - SH wheat (shw), bread wheat (bw), durum wheat (dw) lines, where geneflow across the populations rarely occurs, whereas the secondary population consisted of the synthetic derivative lines (sd) which were derived by the introgression of shw, bw and dw, respectively. We assumed a SNP marker with two alleles (1 and 2) at locus L with an allele frequency P 1shw, P 2shw in the Ae. tauschii – SH wheat population, and P 1bw , P 2bw in the bread wheat population and P 1sd , P 2sd in the synthetic derivative line population. Then the population specific differential (δD) can be represented as:
$${\delta }_{{\rm{D}}(l)}=|{P}_{1shw}^{(l)}-{P}_{2bw}^{(l)}|=|{P}_{2shw}^{(l)}-{P}_{2bw}^{(l)}|$$
(1)
For calculating population specificity at A and B, let allele frequency P1dw, P2dw for durum wheat population, the population specific differential (δA or B) can be represented as:
$${\delta }_{{\rm{A}}or{\rm{B}}(l)}=|{P}_{1dw}^{(l)}-{P}_{2bw}^{(l)}|=|{P}_{2dw}^{(l)}-{P}_{2bw}^{(l)}|$$
(2)
For calculation of D' contribution, the minimum population specific differential (δ D(l)) was set to greater than 0.30 as 80% of the markers had the differential less than 0.3082. Using the allele frequencies, the least-square estimation of the introgression proportion (ϕ), as a measure of the genetic contribution for D' at each locus l 83,84 can be expressed as
$${\varphi }_{l}=\frac{{\sum }_{i=1}^{2}({P}_{shw}^{(l)}-{P}_{bw}^{(l)})({P}_{sd}^{(l)}-{P}_{bw}^{(l)})/{P}_{sd}^{(l)}}{{\sum }_{i=1}^{2}{({P}_{shw}^{(l)}-{P}_{bw}^{(l)})}^{2}/{P}_{sd}^{(l)}}$$
(3)
and overall L loci:
$$\varphi =\frac{{\sum }_{l=1}^{L}{\sum }_{i=1}^{2}({P}_{shw}^{(l)}-{P}_{bw}^{(l)})({P}_{sd}^{(l)}-{P}_{bw}^{(l)})/{P}_{sd}^{(l)}}{{\sum }_{l=1}^{L}{\sum }_{i=1}^{2}{({P}_{shw}^{(l)}-{P}_{bw}^{(l)})}^{2}/{P}_{sd}^{(l)}}$$
(4)
Then sampling variance of \(\varphi \) is: \({\rm{V}}(\varphi )=\frac{MSE}{{\sum }_{l=1}^{L}{\sum }_{i=1}^{2}{({P}_{shw}^{(l)}-{P}_{bw}^{(l)})}^{2}\,/{P}_{sd}^{(l)}}\), where \(MSE=\frac{{\sum }_{l=1}^{L}{\sum }_{i=1}^{2}{[({P}_{sd}^{(l)}-{P}_{bw}^{(l)})(\varphi {P}_{shw}^{(l)}-{P}_{bw}^{(l)})]}^{2}\,/{P}_{sd}^{(l)}}{r-L}\) \({\rm{and}}\,r={{\sum }_{l=1}^{L}r}_{l}\) is the total number of alleles at all L loci.
The D' contribution at each locus was plotted across the wheat chromosome to determine if the contribution is disproportionate in different segments of the genome. The proportion of contribution from the parental populations, D and D', were calculated using the admixture 1.3 program that employs the maximum likelihood approach85,86 under supervised learning mode where reference individuals are members of the SH and bread wheat populations and leaving the synthetic derivative lines as unknown degree of contribution from the two parental populations.
Theoretical contribution estimation
The theoretical contribution of D' to synthetic derivative lines was calculated using the available pedigree information, where pedigrees were extended at multiple levels and were traced back to the first cross between the SH wheat and bread wheat using the International Wheat Information System (IWIS) version 2 (https://www.cimmyt.org/funder_partner/international-wheat-improvement-network-iwin/). IWIS, curated by CIMMYT, has a collection of pedigree and phenotypic data recorded since 1976 to date. Using a recursive approach using the pedigree information, we calculated the theoretical contribution of D'. The φ I , the D' contribution to individual i can be estimated with D contribution to first parent (female), abbreviated as φ P1, and second parent (male), φ P2, with the formula:
$${\phi }_{I}=\frac{1}{2}({\phi }_{P1}+{\phi }_{P2})\times 100$$
(5)
As SH wheat has the entire D' genome derived from Ae. tauschii, the φ I is considered as 100. For bread wheat with no SH wheat in its pedigree, the φ I is considered as 0. The different accessions of Ae. tauschii were assumed to have equal amounts of contribution. The theoretical and marker-estimated contribution of D' was compared across all included synthetic derivative lines.
Average genetic contribution to international nurseries and released cultivars
To estimate the impact of SH wheat in the CIMMYT spring bread wheat breeding program the average theoretical contribution of D' was estimated in two international yield trials, which were annually disseminated to 100–400 national partners worldwide as part of the CIMMYT International Wheat Improvement Network (IWIN). The entries of the Elite Spring Wheat Yield Trial (ESWYT) targeted to irrigated wheat growing areas and the Semi-Arid Wheat Yield Trial (SAWYT) targeted to rain-fed areas from 19 years (1997 to 2016) were used (Table 2). The entries included in these two yield trials are considered to be the best CIMMYT breeding lines for their target environment testing and up-take as cultivars in the developing world. The average contribution of D' in synthetic derivative lines released as cultivars worldwide between 2003–2017 were also analyzed (Table 2, Supplementary Table S2). All calculations and plotting, otherwise specified above, were performed in R87.
Data Availability
The data is available at CIMMYT Dataverse (http://hdl.handle.net/11529/10548269).
References
- 1.
Dubcovsky, J. & Dvorak, J. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science. 316, 1862–1866 (2007).
CAS PubMed PubMed Central Article Google Scholar
- 2.
Salamini, F., Ozkan, H., Brandolini, A., Schäfer-Pregl, R. & Martin, W. Genetics and geography of wild cereal domestication in the near east. Nat. Rev. Genet. 3, 429–441 (2002).
CAS PubMed Article Google Scholar
- 3.
Salse, J. et al. New insights into the origin of the B genome of hexaploid wheat: evolutionary relationships at the SPA genomic region with the S genome of the diploid relative Aegilops speltoides. BMC Genomics 9, 555, https://doi.org/10.1186/1471-2164-9-555 (2008).
CAS PubMed PubMed Central Article Google Scholar
- 4.
Dvorak, J., Akhunov, E. D., Akhunov, A. R., Deal, K. R. & Luo, M. C. Molecular characterization of a diagnostic DNA marker for domesticated tetraploid wheat provides evidence for gene flow from wild tetraploid wheat to hexaploid wheat. Mol. Biol. Evol. 23, 1386–1396 (2006).
CAS PubMed Article Google Scholar
- 5.
International Wheat Genome Sequencing Consortium, A chromosome-based draft sequence of the hexaploid bread wheat genome. Science 345, 1251788 (2014).
- 6.
Sandve, S. R. et al. Chloroplast phylogeny of Triticum/Aegilops species is not incongruent with an ancient homoploid hybrid origin of the ancestor of the bread wheat D-genome. New Phyto. 208, 9–10 (2015).
Article Google Scholar
- 7.
Cox, T. Deepening the wheat gene pool. J. Crop Prod. 1, 145–168 (1997).
Article Google Scholar
- 8.
Varshney, R. K., Nayak, S. N., May, G. D. & Jackson, S. A. Next-generation sequencing technologies and their implications for crop genetics and breeding. Trends Biotechnol. 27, 522–530 (2009).
CAS PubMed Article Google Scholar
- 9.
Davey, J. W. et al. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet. 12, 499–510 (2011).
ADS CAS PubMed Article Google Scholar
- 10.
Cavanagh, C. R. et al. Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc. Natl. Acad. Sci. 110, 8057–8062 (2013).
ADS CAS PubMed Article Google Scholar
- 11.
Dreisigacker, S. et al. Genetic structure of the CIMMYT international yield trial targeted to irrigated environments. Mol. Breed. 29, 529–541 (2012).
Article Google Scholar
- 12.
Sehgal, D. et al. Identification of genomic regions for grain yield and yield stability and their epistatic interactions. Sci. Reports. 7, 41578, https://doi.org/10.1038/srep41578 (2017).
ADS CAS Article Google Scholar
- 13.
Alipour, H. et al. Genotyping-by-sequencing (GBS) revealed molecular genetic diversity of Iranian wheat landraces and cultivars. Front. Plant Sci. 8, 1293, https://doi.org/10.3389/fpls.2017.01293 (2017).
PubMed PubMed Central Article Google Scholar
- 14.
Iehisa, J. C. M. et al. Genome-wide marker development for the wheat D genome based on single nucleotide polymorphisms identified from transcripts in the wild wheat progenitor Aegilops tauschii. Theor. Appl. Genet. 127, 261–271 (2014).
CAS PubMed Article Google Scholar
- 15.
Allen, A. M. et al. Discovery and development of exome-based, co-dominant single nucleotide polymorphism markers in hexaploid wheat (Triticum aestivum L.). Plant Biotechnol. J. 11, 279–295 (2013).
CAS PubMed Article Google Scholar
- 16.
Caldwell, K. S. et al. Sequence polymorphism in polyploid wheat and their D-genome diploid ancestor. Genetics. 167, 941–947 (2004).
CAS PubMed PubMed Central Article Google Scholar
- 17.
Jafarzadeh, J. et al. Breeding value of primary synthetic wheat genotypes for grain yield. PLoS ONE 11, e0162860, https://doi.org/10.1371/journal.pone.0162860 (2016).
CAS PubMed PubMed Central Article Google Scholar
- 18.
Cooper, J. K. et al. Increasing hard winter wheat yield potential via synthetic wheat: I. path-coefficient analysis of yield and its components. Crop Sci. 52, 2014–2022 (2012).
Article Google Scholar
- 19.
Ogbonnaya, F. C. et al. Yield of synthetic backcross-derived lines in rainfed environments of Australia. Euphytica 157, 321–336 (2007).
Article Google Scholar
- 20.
Sehgal, D. et al. Exploring and mobilizing the gene bank biodiversity for wheat improvement. PLoS ONE. 10, e0132112, https://doi.org/10.1371/journal.pone.0132112 (2015).
CAS PubMed PubMed Central Article Google Scholar
- 21.
Young, N. D. & Tanksley, S. D. RFLP analysis of the size of chromosomal segments retained around the Tm-2 locus of tomato during backcross breeding. Theor. Appl. Genet. 77, 353–359 (1989).
CAS PubMed Article Google Scholar
- 22.
Haudry, A. et al. Grinding up wheat: a massive loss of nucleotide diversity since domestication. Mol. Biol. Evol. 24, 1506–1517 (2007).
CAS PubMed Article Google Scholar
- 23.
Dreisigacker, S., Kishii, M., Lage, J. & Warburton, M. Use of synthetic hexaploid wheat to increase diversity for CIMMYT bread wheat improvement. Aust. J. Agric. Res. 59, 413–420 (2008).
Article Google Scholar
- 24.
Li, J., Wan, H. S. & Yang, W. Y. Synthetic hexaploid wheat enhances variation and adaptive evolution of bread wheat in breeding processes. J. Syst. Evol. 52, 735–742 (2014).
Article Google Scholar
- 25.
Bhatta, M., Morgounov, A., Belamkar, V., Poland, J. & Baenziger, P. S. Unlocking the novel genetic diversity and population structure of Synthetic Hexaploid wheat. BMC Genomics 19, 591, https://doi.org/10.1186/s12864-018-4969-2 (2018).
PubMed PubMed Central Article Google Scholar
- 26.
Lage, J., Warburton, M. L., Crossa, J., Skovmand, B. & Andersen, S. B. Assessment of genetic diversity in synthetic hexaploid wheats and their Triticum dicoccum and Aegilops tauschii parents using AFLPs and agronomic traits. Euphytica 134, 305–317 (2003).
CAS Article Google Scholar
- 27.
Zhang, P. et al. Quantifying novel sequence variation and selective advantage in synthetic hexaploid wheats and their backcross-derived lines using SSR markers. Mol. Breed. 15, 1–10 (2005).
Article Google Scholar
- 28.
Gill, B. S. et al. Evaluation of Aegilops species for resistance to wheat powdery mildew, wheat leaf rust, Hessian fly, and greenbug. Plant Dis. 69, 314–316 (1985).
Google Scholar
- 29.
Kihara, H. Discovery of the DD analyser, one of the ancestors of Triticum vulgare. Agric. Hort. 19, 889–890 (1944).
Google Scholar
- 30.
McFadden, E. S. & Sears, E. R. The artificial synthesis of Triticum spelta. Rec. Genet. Soc. Am. 13, 26–27 (1944).
Google Scholar
- 31.
McFadden, E. S. & Sears, E. R. The origin of Triticum spelta and its free-threshing hexaploid relatives. J. Hered. 37, 81–89 (1946).
PubMed Article Google Scholar
- 32.
Mujeeb-Kazi, A., Rosas, V. & Roldan, S. Conservation of the genetic variation of Triticum tauschii (Coss.) Schmalh. (Aegilops squarrosa auct. non L.) in synthetic hexaploid wheats (T. turgidum L. s.lat. X T. tauschii; 2 n = 6x = 42, AABBDD) and its potential utilization for wheat improvement. Gen. Res. Crop Evol. 43, 129–134 (1996).
Article Google Scholar
- 33.
van Ginkel, M. & Ogbonnaya, F. Novel genetic diversity from synthetic wheats in breeding cultivars for changing production conditions. F. Crop. Res. 104, 86–94 (2007).
Article Google Scholar
- 34.
Villareal, R. L., Mujeeb Kazi, A., Del Toro, E., Crossa, J. & Rajaram, S. Agronomic variability in selected Triticum turgidum x T. tauschii synthetic hexaploid wheats. J. Agron. Crop Sci. 173, 307–317 (1994).
Article Google Scholar
- 35.
Villareal, R. L., Mujeeb Kazi, A., Rajaram, S. & Del Toro, E. Morphological variability in some synthetic hexaploid wheats derived from Triticum turgidum x T. tauschii. J. Genet. Breed. 48, 7–15 (1994).
Google Scholar
- 36.
Reynolds, M.P., Skovmand, B., Trethowan, R. & Pfeiffer, W. Evaluating a conceptual model for drought tolerance. In: Ribaut, J. M. (Ed.), Using Molecular Markers to Improve Drought tolerance, 49–53 (CIMMYT, Mexico, D.F., 1999).
- 37.
Calderini, D. F. & Reynolds, M. P. Changes in grain weight as a consequence of de-graining treatments at pre- and post-anthesis in synthetic hexaploid lines of wheat (Triticum durum x T. tauschii). Aust. J. Plant Phys. 27, 183–191 (2000).
Google Scholar
- 38.
Blanco, I. A., Rajaram, S., Kronstad, W. E. & Reynolds, M. P. Physiological performance of synthetic hexaploid wheat-derived populations. Crop Sci. 40, 1257–1263 (2000).
Article Google Scholar
- 39.
Ogbonnaya, F. C. et al. Yield of synthetic backcross-derived lines in rainfed environments of Australia. Euphytica 157, 321–336 (2007).
Article Google Scholar
- 40.
Lopes, M. S. & Reynolds, M. P. Drought adaptive traits and wide adaptation in elite lines derived from resynthesized hexaploid wheat. Crop Sci. 51, 1617–1626 (2011).
Article Google Scholar
- 41.
Ogbonnaya, F. C. et al. Synthetic hexaploids: harnessing species of the primary gene pool for wheat improvement. Plant Breed. Rev. 37, 35–122 (2013).
Article Google Scholar
- 42.
Schachtman, D. P., Lagudah, E. S. & Munns, R. The expression of salt tolerance from Triticum tauschii in hexaploid wheat. Theor. Appl. Genet. 84, 714–719 (1992).
CAS PubMed Article Google Scholar
- 43.
Villareal, R. L., Sayre, K., Banuelos, O. & Mujeeb-Kazi, A. Registration of four synthetic hexaploid wheat (Triticum turgidum/Aegilops tauschii) germplasm lines tolerant to waterlogging. Crop Sci. 41, 274 (2001).
Article Google Scholar
- 44.
Das, M. K., Bai, G., Mujeeb-Kazi, A. & Rajaram, S. Genetic diversity among synthetic hexaploid wheat accessions (Triticum aestivum) with resistance to several fungal diseases. Genet. Resour. Crop Evol. 63, 1285–1296 (2016).
CAS Article Google Scholar
- 45.
Morgounov, A. et al. High-yielding winter synthetic hexaploid wheats resistant to multiple diseases and pests. Plant Genet. Resour. 16, 273–278 (2018).
CAS Article Google Scholar
- 46.
Zegeye, H., Rasheed, A., Makdis, F., Badebo, A. & Ogbonnaya, F. C. Genome-wide association mapping for seedling and adult plant resistance to stripe rust in synthetic hexaploid wheat. PLoS ONE. 9, e10559, https://doi.org/10.1371/journal.pone.0105593 (2014).
CAS Article Google Scholar
- 47.
Assefa, S. & Fehrmann, H. Resistance to wheat leaf rust in Aegilops tauschii Coss. and inheritance of resistance in hexaploid wheat. Genet. Res. Crop Evol. 47, 135–140 (2000).
Article Google Scholar
- 48.
Ma, H., Singh, R. P. & Kazi Mujeeb, A. Resistance to stripe rust in Triticum turgidum, T. tauschii and their synthetic hexaploids. Euphytica 82, 117–124 (1995).
Article Google Scholar
- 49.
Marais, G. F., Potgieter, G. F. & Roux, H. S. An assessment of the variation for stem rust resistance in the progeny of a cross involving the Triticum species aestivum, turgidum and tauschii. S. A. J. Plant Soil. 11, 15–19 (1994).
Article Google Scholar
- 50.
Arraiano, L. S., Worland, A. J., Ellerbrook, C. & Brown, J. K. M. Chromosomal location of a gene for resistance to septoria tritici blotch (Mycosphaerella graminicola) in the hexaploid wheat'Synthetic 6x'. Theor. Appl. Genet. 103, 758–764 (2001).
CAS Article Google Scholar
- 51.
Loughman, R., Lagudah, E. S., Trottet, M., Wilson, R. E. & Mathews, A. Septoria nodorum blotch resistance in Aegilops tauschii and its expression in synthetic amphiploids. Aust. J. Agric. Res. 52, 1393–1402 (2001).
Article Google Scholar
- 52.
Mujeeb-Kazi, A., Cano, S., Rosas, V., Cortes, A. & Delgado, R. Registration of five synthetic hexaploid wheat and seven bread wheat lines resistant to wheat spot blotch. Crop Sci. 4, 1653–1654 (2001).
Article Google Scholar
- 53.
Mujeeb-Kazi, A., Delgado, R., Juarez, L. & Cano, S. Scab resistance (Type II: spread) in synthetic hexaploid germplasm. Ann. Wheat News. 47, 118–120 (2001).
Google Scholar
- 54.
Cox, T. S. et al. Resistance to foliar diseases in a collection of Triticum tauschii germplasm. Plant Dis. 76, 1061–1064 (1992).
Article Google Scholar
- 55.
Kong, L. et al. Location of a powdery mildew resistance gene in Am6, an amphidiploid between Triticum durum and Aegilops tauschii, and its utilisation. Acta Phytophyl. Sinica 26, 116–120 (1999).
CAS Google Scholar
- 56.
Villareal, R. L., Mujeeb Kazi, A., Fuentes Davila, G., Rajaram, S. & Del Toro, E. Resistance to Karnal bunt (Tilletia indica Mitra) in synthetic hexaploid wheats derived from Triticum turgidum x T. tauschii. Plant Breed. 112, 63–69 (1994).
Article Google Scholar
- 57.
Eastwood, R. F., Lagudah, E. S., Appels, R., Hannah, M. & Kollmorgen, J. F. Triticum tauschii: a novel source of resistance to cereal cyst nematode (Heterodera avenae). Aust. J. Agric. Res. 42, 69–77 (1991).
Google Scholar
- 58.
Thompson, J. P., Brennan, P. S., Clewett, T. G., Sheedy, J. G. & Seymour, N. P. Progress in breeding wheat for tolerance and resistance to root-lesion nematode (Pratylenchus thornei). Austr. Plant Path. 28, 45–52 (1999).
Article Google Scholar
- 59.
Hollenhorst, M. M. & Joppa, L. R. Chromosomal location of genes for resistance to greenbug in 'Largo' and 'Amigo' wheats. Crop Sci. 23, 91–93 (1983).
Article Google Scholar
- 60.
Tyler, J. M. & Hatchett, J. H. Temperature influence on expression of resistance to Hessian fly (Diptera: Cecidomyiidae) in wheat derived from Triticum tauschii. J. Econ. Ento. 76, 323–326 (1983).
Article Google Scholar
- 61.
William, M., Pena, R. J. & Mujeeb Kazi, A. Seed protein and isozyme variations in Triticum tauschii (Aegilops squarrosa). Theor. Appl. Genet. 87, 257–263 (1993).
CAS PubMed Article Google Scholar
- 62.
Pena, R. J., Zarco Hernandez, J. & Mujeeb Kazi, A. Glutenin subunit compositions and bread-making quality characteristics of synthetic hexaploid wheats derived from Triticum turgidum x Triticum tauschii (coss) Schmal crosses. J. Cereal Sci. 21, 15–23 (1983).
Article Google Scholar
- 63.
Pfluger, L. A. et al. Characterisation of high- and low-molecular weight glutenin subunits associated to the D genome of Aegilops tauschii in a collection of synthetic hexaploid wheats. Theor. Appl. Genet. 103, 1293–1301 (2001).
CAS Article Google Scholar
- 64.
Yang, W. et al. Synthetic hexaploid wheat and its utilization for wheat genetic improvement in China. J. Genet. Genomics. 36, 539–546 pmid:19782955 (2009).
CAS PubMed Article Google Scholar
- 65.
Li, J. et al. Identification of a high-yield introgression locus in Chuanmai 42 inherited from synthetic hexaploid wheat. Acta Agron. Sin. 37, 255–262 (2011).
CAS Google Scholar
- 66.
The International Wheat Genome Sequencing Consortium, Shifting the limits in wheat research and breeding using a fully annotated reference genome, Science, 361, eaar7191, https://doi.org/10.1126/science.aar7191 (2018).
- 67.
Cox, T. S. et al. Comparing two approaches for introgression of germplasm from Aegilops tauschii into common wheat. The Crop J. 5, 355–362 (2017).
Article Google Scholar
- 68.
Velu, G., Ortiz-Monasterio, I., Cakmak, I., Hao, Y. & Singh, R. P. Biofortification strategies to increase grain zinc and iron concentrations in wheat. J. Cereal Sci. 59, 365–372 (2014).
CAS Article Google Scholar
- 69.
Rowland, G. G. & Kerber, E. R. Telocentric mapping in hexaploid wheat of genes for leaf rust resistance and other characters derived from Aegilops squarrosa. Can. J. Genet. Cytol. 16, 137–144 (1974).
Article Google Scholar
- 70.
Singh, R. P., Nelson, J. C. & Sorrells, M. E. Mapping Yr28 and other genes for resistance to stripe rust in wheat. Crop Sci. 40, 1148–1155 (2000).
CAS Article Google Scholar
- 71.
Olson, E. L. et al. Introgression of stem rust resistance genes SrTA10187 and SrTA10171 from Aegilops tauschii to wheat. Theor. Appl. Genet. 126, 2477–2484 (2013).
CAS PubMed Article Google Scholar
- 72.
Pranger, A. L. Synthetic hexaploid wheat contributes favorable alleles for yield and yield components in an advanced backcross winter wheat population. M.S. thesis, Colorado State University, Fort Collins, CO. Available, https://mountainscholar.org/handle/10217/66666 (2011).
- 73.
Dreccer, F. M., Borgognone, G. M., Ogbonnaya, F. C., Trethowan, R. M. & Winter, B. CIMMYT-selected derived synthetic bread wheats for rainfed environments: yield evaluation in Mexico and Australia. Field Crops Res. 100, 218–228 (2007).
Article Google Scholar
- 74.
Rattey, A. & Shorter, R. Evaluation of CIMMYT conventional and synthetic spring wheat germplasm in rainfed sub-tropical environments. I. Grain yield. Field Crops Res. 118, 273–281 (2010).
Article Google Scholar
- 75.
Martin, S. H. & Jiggins, C. D. Interpreting the genomic landscape of introgression. Cur. Opin. Genet. Dev. 47, 69–74 (2017).
CAS Article Google Scholar
- 76.
Campbell, L. G. & Snow, A. A. Competition alters life history and increases the relative fecundity of crop–wild radish hybrids (Raphanus spp.). New Phyto. 173, 648–660 (2007).
Article Google Scholar
- 77.
Kost, M. A., Alexander, H. M., Jason Emry, D. & Mercer, K. L. Life history traits and phenotypic selection among sunflower crop–wild hybrids and their wild counterpart: Implications for crop allele introgression. Evol. Appl. 8, 510–524 (2015).
PubMed PubMed Central Article Google Scholar
- 78.
Corbi, J., Baack, E. J. & Burke, J. M. Genome-wide analysis of allele frequency change in sunflower crop–wild hybrid populations evolving under natural conditions. Mol. Ecol. 27, 233–247 (2018).
CAS PubMed Article Google Scholar
- 79.
Trethowan, R. & Mujeeb-Kazi, A. Novel germplasm resources for improving environmental stress tolerance of hexaploid wheat. Crop Sci. 48, 1255–1265 (2008).
Article Google Scholar
- 80.
Chakraborty, R., Kamboh, M. I., Nwankwo, M. & Ferrell, R. E. Caucasian genes in American blacks: new data. Am. J. Hum. Genet. 50, 145–155 (1992).
CAS PubMed PubMed Central Google Scholar
- 81.
Stephens, J. C., Briscoe, D. & O'Brien, S. J. Mapping by admixture linkage disequilibrium in human populations: limits and guidelines. Am. J. Hum. Genet. 55, 809–824 (1994).
CAS PubMed PubMed Central Google Scholar
- 82.
Shriver, M. D. et al. Ethnic-affiliation estimation by use of population-specific DNA markers. Am. J. Hum. Genet. 60, 957–964 (1997).
CAS PubMed PubMed Central Google Scholar
- 83.
Long, J. C. The genetic structure of admixed populations. Genetics 127, 417–428 (1991).
CAS PubMed PubMed Central Google Scholar
- 84.
Long, J. C. & Smouse, P. E. Inter tribal geneflow between the Ye'cuana and Yanomama: genetic analysis of an admixed village. Am. J. Phys. Anthropo. l61, 411–422 (1983).
Article Google Scholar
- 85.
Alexander, D. H. & Lange, K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinfo. 12, 246, https://doi.org/10.1186/1471-2105-12-246 (2011).
Article Google Scholar
- 86.
Alexander, D., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009).
CAS PubMed PubMed Central Article Google Scholar
- 87.
R Core Team R, A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. available at, http://www.R-project.org/ (2018).
Download references
Acknowledgements
This study was supported by CRP-WHEAT and SEED of Discovery project (MasAgro). We acknowledge the contribution of our worldwide partners who participated in evaluation of CIMMYT international wheat yield trials.
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
Reprints and Permissions
About this article
Cite this article
Rosyara, U., Kishii, M., Payne, T. et al. Genetic Contribution of Synthetic Hexaploid Wheat to CIMMYT's Spring Bread Wheat Breeding Germplasm. Sci Rep 9, 12355 (2019). https://doi.org/10.1038/s41598-019-47936-5
Download citation
-
Received:
-
Accepted:
-
Published:
-
DOI : https://doi.org/10.1038/s41598-019-47936-5
Further reading
-
How the pan-genome is changing crop genomics and improvement
Genome Biology (2021)
-
Evaluation of genetic structure in European wheat cultivars and advanced breeding lines using high-density genotyping-by-sequencing approach
BMC Genomics (2021)
-
RNA-Seq-based DNA marker analysis of the genetics and molecular evolution of Triticeae species
Functional & Integrative Genomics (2021)
-
Biofortification and bioavailability of Zn, Fe and Se in wheat: present status and future prospects
Theoretical and Applied Genetics (2021)
-
A GBS-based GWAS analysis of adaptability and yield traits in bread wheat (Triticum aestivum L.)
Journal of Applied Genetics (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Picture Difference in Height Between Hybridized and Ancient Wheat
Source: https://www.nature.com/articles/s41598-019-47936-5
0 Response to "Picture Difference in Height Between Hybridized and Ancient Wheat"
Post a Comment